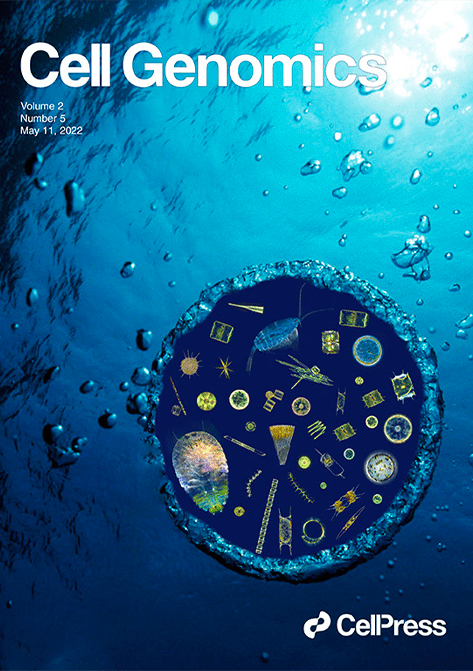For more than 10 years now, the plankton samples collected from across the globe by the Tara Oceans expeditions have been continuously delivering new discoveries on this vital ecosystem for the planet and humankind. For this grand scientific adventure, Genoscope sequences and analyzes the DNA found within the water samples collected from more than 150 oceanic sites and at a variety of water depths. The method used by the laboratory, called “metagenomics” , has shown its potential by delivering the DNA sequences of all of the organisms present in each water sample.
Making up a large part of the many species present in plankton, eukaryotes are complex, nucleated organisms that live mainly near the ocean surface. They comprise unicellular organisms, microscopic animals and photosynthetic algae among others. By contributing greatly to the flux of matter, energy and oxygen in the ocean, eukaryotic plankton plays a key role in the stability of the entire marine ecosystem.
Compared to those of prokaryotes, the genomes of eukaryotes are larger, more complex, divided into chromosomes and marked by numerous non-coding and repetitive sequences. Because of that complexity, reconstructing individual eukaryote genomes from the global metagenome derived from oceanic water samples had remained out of science’s reach. Until now. Indeed, an international team piloted by the CNRS and Genoscope (CEA/CNRS/University of Évry-Paris Saclay) has finally touched that goal. Benefiting from the partial exclusion of prokaryotes via the size fractionation methodology available on the schooner Tara, the researchers worked on the massive amount of sequencing data from close to 800 metagenomics samples, representing 280 billion short DNA sequences. Their unprecedented results earned the cover of the 11 May 2022 edition of Cell Genomics.
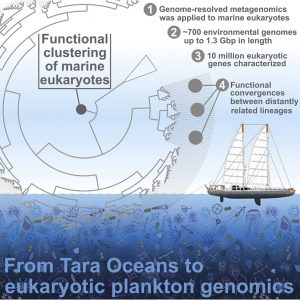 The work reconstructed, at least partially, close to 700 planktonic eukaryote species genomes, and in so doing, established the most precise representation of the diversity of those species available today. The researchers were able to place the organisms into five groups sharing biological functions, one comprising multicellular species and the four others unicellular species. Surprisingly, two groups brought together taxonomically-distant organisms, demonstrating functional convergence (developing the same adaptive strategies) among species sharing a same oceanic ecosystem for millions of years. For example, similar sugar transport and metabolism functions were found in a diverse group of large eukaryotes, perhaps associated with a shared prey.
The work reconstructed, at least partially, close to 700 planktonic eukaryote species genomes, and in so doing, established the most precise representation of the diversity of those species available today. The researchers were able to place the organisms into five groups sharing biological functions, one comprising multicellular species and the four others unicellular species. Surprisingly, two groups brought together taxonomically-distant organisms, demonstrating functional convergence (developing the same adaptive strategies) among species sharing a same oceanic ecosystem for millions of years. For example, similar sugar transport and metabolism functions were found in a diverse group of large eukaryotes, perhaps associated with a shared prey.