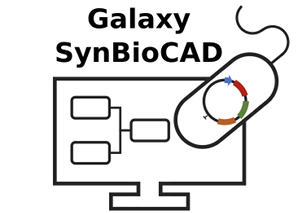
 On Wednesday, 26 November, Joan Hérisson, BioRetroSynth research engineer within the Genomics Metabolics Unit (CEA/CNRS/University of Evry Paris-Saclay), presented the Galaxy-SynBioCAD synthetic biology portal, at the International Workshop on Bio-Design Automation (IWBDA), held in Paris in parallel with the renowned international synthetic biology competition iGEM.
On Wednesday, 26 November, Joan Hérisson, BioRetroSynth research engineer within the Genomics Metabolics Unit (CEA/CNRS/University of Evry Paris-Saclay), presented the Galaxy-SynBioCAD synthetic biology portal, at the International Workshop on Bio-Design Automation (IWBDA), held in Paris in parallel with the renowned international synthetic biology competition iGEM.
Widening the scope of bioproduction
Increasing the number of compounds able to be produced via biological pathways is a major issue for the transformation of synthetic chemistry into a more sustainable bioindustry. But, is it possible to construct a metabolic pathway for any given compound of interest? Can a microorganism by programmed to synthesize it?
Conceived by J.-L. Faulon’s team (Institut MICALIS, INRAE)), which benefits from the presence of several Genopole bioinformaticians, the online portal Galaxy-SynBioCAD can answer those questions.
By uniting a range of synthetic biology tools, guaranteeing their compatibility with the use of standardized languages such as SBML and SBOL, and tying them all together in the workflow platform Galaxy, Galaxy-SynBioCAD is able to:
- carry out the complete process of design and engineering for metabolic pathways;
- ascertain what enzymes are needed for a given metabolic pathway;
- determine the assembly of DNA sequences (coding for the enzymes);
- and generate handler scripts for the assembly of plasmids and the production of compounds of interest.
Galaxy-SynBioCAD: all design and engineering steps in just a few clicks
In concrete terms, Galaxy-SynBioCAD delivers a plasmid containing the genes that code for an enzyme able to catalyze the biochemical reactions needed to synthesize a compound of interest. To validate their tool, the authors made it available to other teams and tested it on literature and expert-validated metabolic pathways.
For example, deploying the tool at four different locations, they demonstrated reproducibility by constructing a collection of Escherichia coli strains producing lycopene, an easily identified red pigment. Furthermore, the team benchmarked the tool against 60 molecules previously described in the literature, reporting that in 83% of cases, the literature or expert selected pathways were found in the first 10 pathways generated by Galaxy-SynBioCAD for a same targeted compound in a same bacterial chassis. To classify the validity of synthesis pathways proposed by Galaxy-SynBioCAD, the authors developed a machine learning scoring process trained on a database of nearly 8000 metabolic pathways.
An article published in Nature Communications on 29 August 2022 presented the open-source Galaxy-SynBioCAD portal, for which the Galaxy platform created a new “Synthetic Biology” category specifically.
The work by this Genoscope team in biological engineering, aimed at enabling the bioproduction of a vast range of chemical compounds, contributes to the construction of a bioeconomy sector at Genopole.