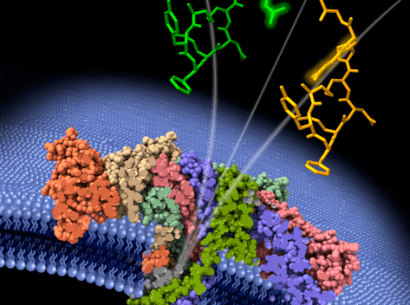
Aerolysin is a toxin produced naturally by Pseudomonas aeruginosa. It forms a nanopore, i.e., an opening measuring only a few nanometers in diameter, in cells during infection. That aerolysin nanopore can however also be used as an ultra-sensitive, selective and specific sensor in a number of biotech settings (Cressiot et al., 2019, ACS Sensors). In their work, the Lambe team showed that under controlled experimental conditions, the properties of aerolysin nanopores could be harnessed to identify 13 of the 20 natural amino acids.
A specific electronic signal during nanopore passage
To achieve that feat, the team associated each amino acid with a positively-charged peptide. Under the influence of an electric field, the amino acid with its peptide enters the nanopore where it is temporarily confined. That confinement blocks the electric current, with each amino acid generating a specific electrical signature. The nanopore is even sensitive enough to detect post-translational modifications, that is, chemical changes that amino acids undergo after protein synthesis. Those modifications are important because they regulate the cellular activities of proteins.
Why characterize the proteome?
The work by the Lambe team is a first step toward the sequencing of proteins, which ensure cellular function.
Sequencing the proteome of a cell or a biological fluid is a major issue in healthcare. In the same way that the genome designates the total genetic information of an organism, the proteome provides the total proteinaceous information of a cell, and the chemical modifications those proteins undergo. Thus, the proteome is the organism’s functional motor.
In contrast to the genome, the proteome varies with the particular functions of a given cell. Importantly, it also bears the marks of cellular dysfunction, notably in disease. For that reason, finely characterizing the proteome may make it possible to diagnose serious diseases such as cancers earlier in their course, prevent them from recurring or develop personalized treatments for them.
The team has its sights set on improving instrumentation and engineering the nanopores to enable the identification of the seven remaining amino acids. Once accomplished, the team will have opened a potential path toward protein sequencing via aerolysin nanopores for unique molecules.
This recent work, published in Nature Biotechnology (2020) follows upon a first study published in Nature Communications (2018) on the detection of homopeptides and the determination of their sequence size using the aerolysin nanopore. The Lambe project is carried out in partnership with Lariboisière Hospital, the start-up DreamPore, the University of Illinois and the University of Freiburg.