The Sysfate laboratory was created within the Genomics Metabolics unit at Genoscope through a Genopole Atige grant. The lab’s director, Marco Mendoza, and his team have developed MULTILAYER, a method for furnishing spatial digital images of activity in biological tissues. The team took inspiration from image analysis and admixed their expertise in “omics” (genomics, transcriptomics, etc.) to create the tool.
MULTILAYER thus reveals the functional structuration of an organ or tissue.
The method will be of particular interest for developing solutions in molecular diagnostics.
The function of a biological tissue or organ is determined by the activity of its constituent cells, or more precisely by the expression or non-expression of their genes.
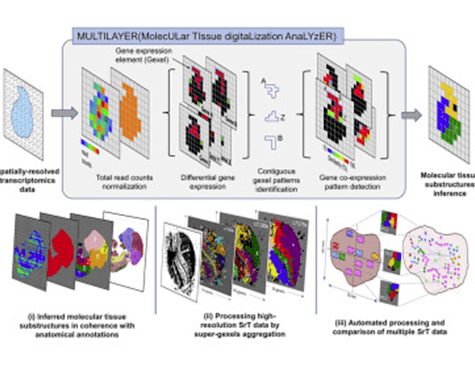
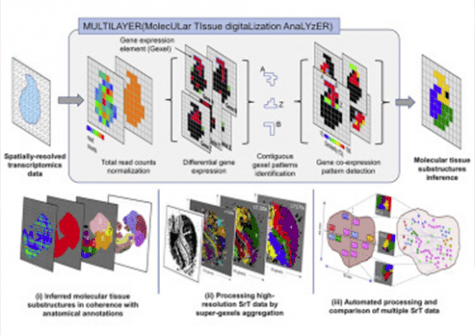
Spatial transcriptomics* is a technique for analyzing the expression levels of a large group of genes simultaneously at the scale of a tissue or organ. From a histological section, the technique furnishes a two-dimensional image of gene expression therein, presented as points distributed evenly over the tissue. The functional map of the tissue can represent a thousand genes and several thousand positions, resulting together in millions of analyzable sequences. Such a quantity of data requires specific statistical and bioinformatics methods. To improve upon current methods, Marco Mendoza and his team at the Sysfate lab (Genoscope) have developed MULTILAYER and described its performance in an article published on 7 May 2021 in Cell Systems.
MULTILAYER analyzes a biological section and assigns to each spatial point in the tissue a measurement, called a gexel by the team for its analogousness with the pixels that make up a digital image. A gexel represents the expression of a thousand genes at a point in the tissue. Compared to conventional statistical methods, the novel nature of MULTILAYER is to be found in its analysis of gexels not as independent entities but as a continuity: adjacent gexels showing similar genetic expression profiles have weight in the analysis of any one gexel. Indeed, from a biological standpoint, contiguous cells should have a greater chance of a shared destiny and thus a shared genetic programming. The Sysfate team’s algorithm also mines known information centralized in databases, notably for gene co-expression, which can potentially indicate groups of genes associated with a given function.
MULTILAYER thus “digitizes” tissue into biologically pertinent substructures.
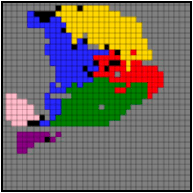
In their article, the Sysfate researchers demonstrated MULTILAYER’s ability to identify functional zones from spatial transcriptomics data derived not only from heart tissue sections and a range of tumor tissues (below figure) but also from an entire mouse embryo, which the tool was able to stratify into regions corresponding perfectly to known anatomical structures (opposite figure).
MULTILAYER is freely available as an autonomous computer program: https://github.com/SysFate/MULTILAYER