Researchers from two Genopole entities, the laboratory SABNP (Université d’Evry Paris-Saclay) and the startup Synsight, have developed a new drug screening approach built upon computational modeling, the structural analysis of biological molecules and a patented technology for the study of RNA-protein interactions (RPIs) within living cells.
The team aims to enable the exploration of the modulation or inhibition of these interactions for therapeutic purposes, a pathway heretofore hampered by a lack of methodologies, but one holding great promise in certain cancers and other pathologies.
In living systems, bindings between proteins and RNA (e.g., protein-coding messenger RNA, other non-coding RNA with diverse biological roles) are essential phenomena for cell function, and thus essential for the entire organism. The human genome codes for at least 1,000 RNA-binding proteins, many of which participate in gene expression. They sometimes play roles in certain pathologies, such as neurodegenerative disorders and cancers.
A path yet to explore toward novel medicines
RPIs constitute a promising pathway for the discovery of new therapeutics. Currently however, it is an underexplored pathway notably due to two obstacles: on one hand, there is a lack of computational models for in silico screening of compounds able to target the binding pocket where the interaction takes place, and on the other, it is difficult to experimentally detect these interactions in cells.
To address these difficulties, the laboratory SABNP, headed by David Pastré and under the supervision of the University of Évry-Paris Saclay, and the start-up Synsight, founded by Cyril Bauvais, joined forces to develop a therapeutic screening approach exploiting chemical, structural and cellular data.
This approach is three-pronged, called upon:
- Large-scale computational models to assess millions of compounds for their target pocket binding aptitude and integrate their absorption, distribution and toxicity characteristics and other physicochemical properties useful for determining their therapeutic potential.
- SABNP’s structural biology competencies, notably those concerning 3D study of the target binding pocket and nuclear magnetic resonance (NMR) spectroscopy analyses of candidate molecules for it.
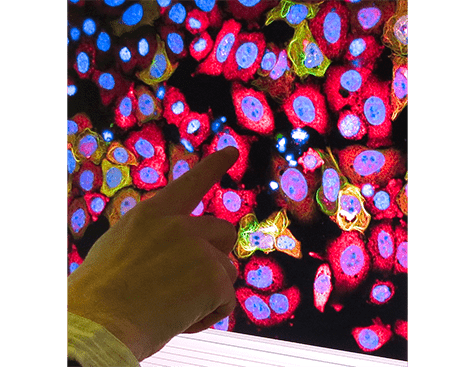
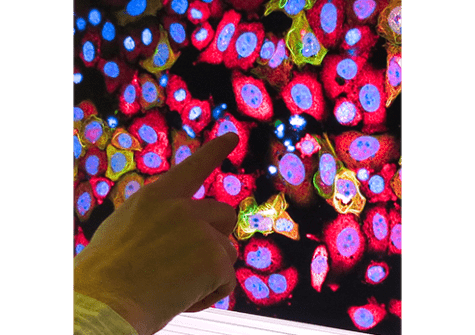
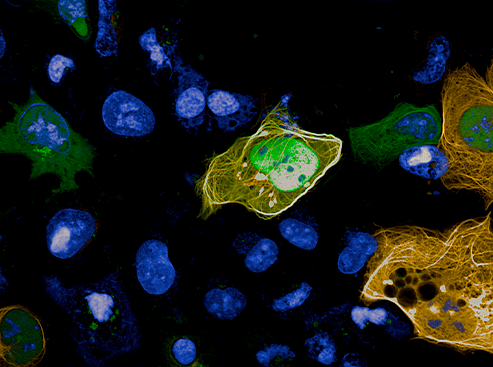
- the SABNP-patented technology MicroTubule bench (MT bench), which uses a naturally occurring structure in cells, the microtubules, to observe protein-protein or RNA-protein interactions in cells (see photo). Associated with a high-content screening system (acquired with Genopole and University of Évry support) and automated imaging technology, the in vivo test platform is able to visualize and quantify these interactions.
Performance demonstrated on YB-1–RNA interaction
The SABNP-Synsight team demonstrated proof of concept for their integrated drug-discovery approach using Y-box binding protein 1 (YB-1), a messenger RNA-binding protein involved in cancer progression and resistance to chemotherapy.
Their study focused on finding YB-1–RNA binding inhibitors within a library of more than seven million compounds and close to five thousand FDA-approved drugs, representing collectively more than 200 million pharmacophores (potential pharmacologically-targetable molecular features) to be evaluated. The paired consideration of in silico binding-potential predictions concerning the YB-1–RNA complex and of criteria for drug development potential (availability, price, purity, etc.) led to the identification of 140 potential inhibitors. Thereafter, computer simulations of bind stability reduced that number to 22..
This impressive, large-scale screening result done in silico thus enabled further screening refinements in ex silico experimental analyses. MT bench cellular testing was implemented in near-natural conditions, using the endogenous messenger RNA present in the cytoplasm of the employed cell lines. And indeed, interference of the YB-1–RNA interaction was detected and measured robustly thanks to the high-resolution HCS imaging and the automated analysis of the thousands of generated images.
With an additional, precise structural analysis of the target binding pocket by NMR spectroscopy the team’s results not only validated the predictive potential of in silico screening but also identified a final 11 compounds that significantly disrupt binding between YB-1 and messenger RNA, in the test tube and in cells.
Among these latter was P1, an FDA-approved polymerase inhibitor, which showed high selectivity and good affinity as concerns the YB-1–RNA interaction.