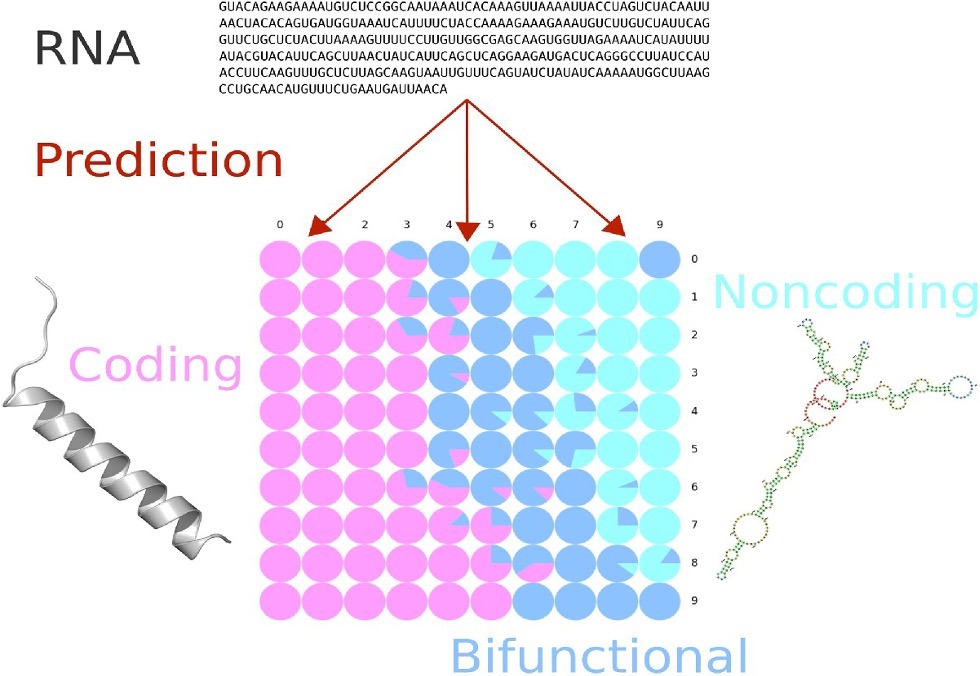
The IBISC laboratory (University Evry Paris-Saclay) has developed IRSOM2, an unprecedented bioinformatics tool for the prediction of bifunctional RNA, a relatively recently discovered class of RNA showing both an ability to code for proteins and an ability to perform biological roles itself. IRSOM2 uses neural networks and self-organizing maps to establish three classes: protein-coding, protein-non-coding and potentially bifunctional RNA. Freely available and easy to use, IRSOM2 furnishes biologists with candidates to study and thus contributes to increasing scientific knowledge in the emerging field of bifunctional RNA.
Over the last two decades, scientists have demonstrated the central role of non-coding RNA in both normal cell processes and certain diseases, notably cancer. Arising from genome regions unassociated with protein coding and thus previously considered as “junk” DNA, these non-coding RNA have been shown to provide key cellular functions like gene expression regulation themselves, instead of producing a protein for the task. More recently, yet another class has emerged, called bifunctional RNA. These latter show both coding and non-coding functions, meaning they are able to both code for the synthesis of a biologically active protein and directly provide regulatory functions themselves. This double-function nature varies according to, for example, environmental parameters, the organism’s development stage, or pathological processes.
Back in 2019, the RNA bioinformatics group headed by Fariza TAHI, within the AROB@S team at the Genopole lab IBISC (University Evry Paris-Saclay), developed IRSOM, This innovative bioinformatics tool identifies coding and non-coding RNA according to open reading frame sequences (« Open Reading Frame » a span of DNA potentially able to be expressed into a protein), k-mers (a bioinformatics measure of sequences of length k), the frequencies of certain nucleotides, and several other parameters. IRSOM functions on a neural network model and uses self-organizing maps to furnish biologists with an easily-interpretable graphical presentation of results. IRSOM also has the particularity of providing a rejection option that identifies ambiguous cases.
To accompany the emergence of bifunctional RNA, Fariza Tahi’s team has now developed IRSOM2. This new tool exploits the original IRSOM’s ability to reject ambiguous cases. Indeed, the research team hypothesized that there was a strong likelihood that these ambiguous cases may show bifunctionality.
The researchers thus validated their strategy and adjusted the probability thresholds for IRSOM2, calling particularly upon the SPENCER database of long RNAs involved in cancer, and the cncRNAdb database of diverse RNAs from a range of organisms. In this manner, the third class of ambiguous RNAs in IRSOM was morphed into one of candidate bifunctional RNAs in IRSOM2 for further study by biologists.
Because knowledge on bifunctional RNA is currently incomplete, IRSOM2 gives teams the ability to autonomously re-train the model on their particular datasets, thus creating their own model.