The bioinformatics group directed by Fariza Tahi within the IBISC laboratory specializes in the identification of RNA sequences and the determination of their structures. That team recently passed a milestone with the development of their C-RCPred model. This latter can predict the 3D organization of multi-RNA complexes, which play important roles in the cell. C-RCPred is the first algorithm to integrate experimental or predicted binding biochemistry data and the specific knowledge on complexes brought to the game by the users themselves. The tool is freely available to the scientific community on the Genopole computer program platform EvryRNA.
RNA is able to pair with other molecules, resulting in RNA complexes that play major roles within cells. Ribosomes, for example, are RNA-protein complexes responsible for the translation of the genetic code into proteins. They thus are a key element of correct cellular function. There are also complexes comprising only RNA molecules, which act as catalysts. In all cases, the 3D structure of the RNA complex is essential to its biological function.
To help biologists better understand RNA complexes, the RNA bioinformatics team at the
Beyond this specificity, C-RCPred also provides:
- a visualization of the predicted structures via a dynamic graphic interface;
- the possibility for biologists to indicate corrections for the structure and re-run the tool with them, and repeatedly so as many times as they wish.
The IBISC team demonstrated C-RCPred’s performance
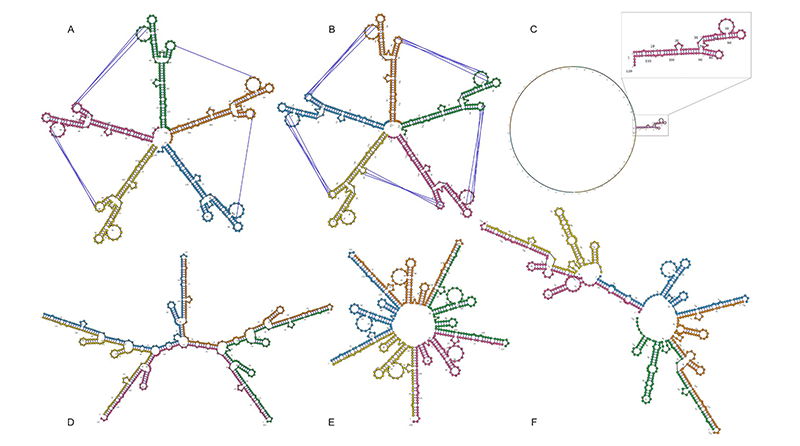
by applying it to the prediction of the secondary structure of a well-characterized (image A in the figure below; PDB code 1FOQ) five-RNA assembly vital to the replication of the bacteriophage φ29. Contrary to other available prediction tools, and with no user constraints applied to the model, C-RCPred was able to precisely determine the secondary structure (prediction B in the figure below) of this long, 597-base-pair complex. RCPred, the team’s initial tool, correctly predicted a branch of the structure (prediction C) whereas the other available tools predicted structures very different from the pentamer’s true form (predictions D, E and F).